Technology
Data Content
The primary data objects in TreeBASE are bibliographic references to published phylogenetic studies, taxon by character data matrices, and phylogenetic trees resulting from the analysis of such data matrices. Information is also available that links data matrices and trees, including types of analyses performed, software used, etc. Wherever possible, taxon labels are mapped to uBio's name services and NCBI's taxonomy.
Implementation Technologies
- Database: PostgreSQL 8.4.2
- Programming language: Java (web-application), Perl (data migration and maintenance)
- Database ORM: Hibernate
- Web-application framework: Spring
- Submission file parsing: Mesquite
- Tree Visualization: PhyloWidget
Source Code
Code developed for the TreeBASE project is part of the TreeBASE GitHub organization. The source code for the web application and the database can be downloaded from the git repository under a BSD license. Documentation on installing and running TreeBASE can be found at the TreeBASE wiki.
Architecture
TreeBASE has a tiered Java-based architecture using the Hibernate and Spring frameworks built on a PostgreSQL database. The following schematic illustrates the content and features of this software stack:
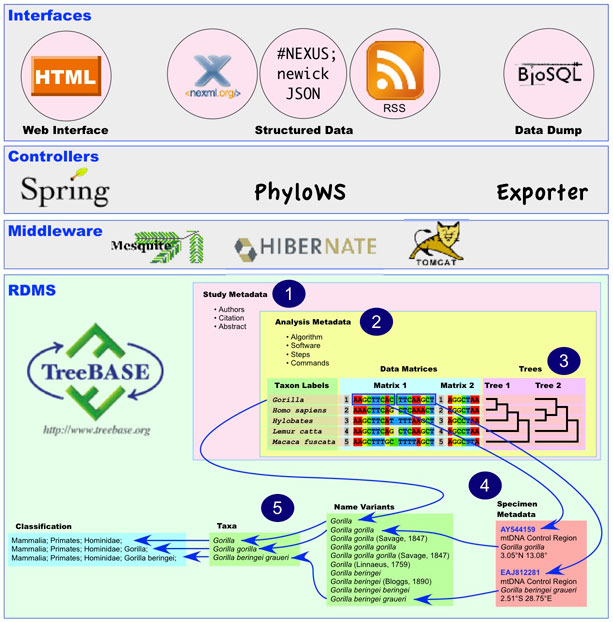
- TreeBASE stores alignments, character matrices, and trees used for research that is published in peer-reviewed journals and books.
- Each study has one or more analyses; each analysis has one or more steps; each step associates matrices and trees with algorithms and software.
- Each row of sequence alignments or coded characters has a taxon label that maps to leaf nodes on associated trees. Trees are hashed to allow topological querying.
- Each row in a character matrix can be subdivided into one or more row segments; each row segment can have associated specimen, tissue, or gene sequence metadata.
- Names in the rows of a matrix and in row segment metadata can independently map to a dictionary of name variants, which maps to a taxonomy. This taxonomy will be mapped to a classification tree in the next release.




